Generative Models for Learning Underlying Geometry of Neuroimaging Data
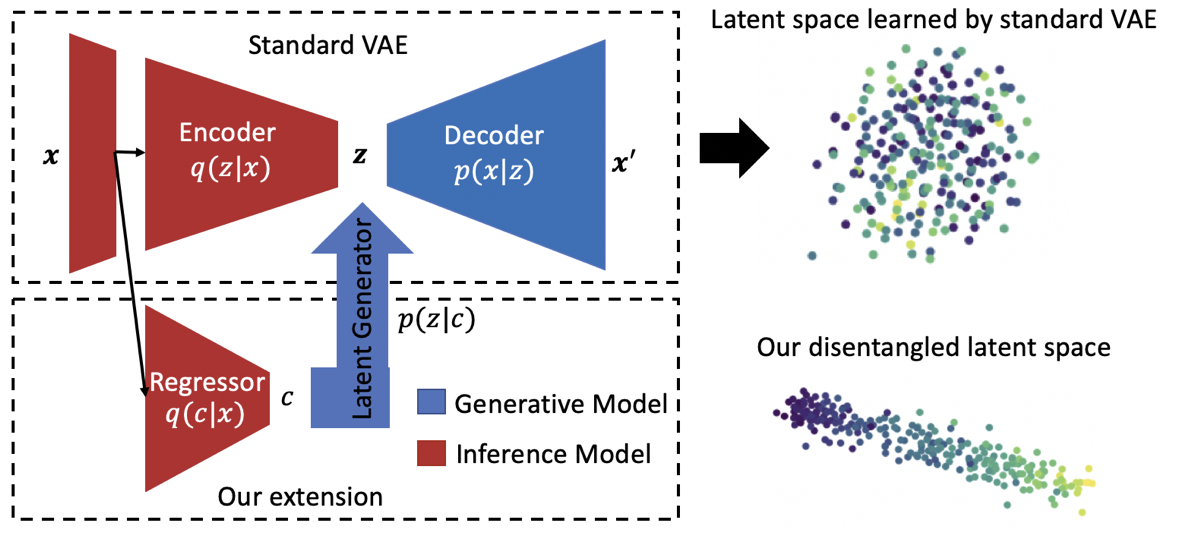
Training deep neural networks on neuroimaging data can be very challenging as studies generally only contain a limited number of samples and the samples do not fully capture the heterogeneity of the population. The risk of biasing the prediction process is further increased by so called confounding factors. For example, age can be a confounding factor in case the disease causes accelerated aging and the given data sets consists of patients that are significantly older than the healthy controls. One way to account for these issues in the training process is to first reduce the complex information captured by neuroimages to a low-dimensional encoding (a.k.a. latent space) before performing the analysis of interest. The low-dimension encoding can explicitly model confounding factors, be used to generate high-quality MRI scans augmenting the training data set, or ease detection of biomarkers specific to a disease.
Motivated by recent advances in deep neural networks, we are exploring novel generative models (e.g., variational autoencoders, style GAN, diffusion models, flow models) to create latent space representations that allow us to perform tasks such as data synthesis, data editing, and feature disentanglement. The resulting models can adapt to both supervised, self-supervised, or unsupervised applications based on both structural and functional MRI data. Examples are the encoding of normal brain aging and deviations caused by neurogenerative diseases [1-4], to discover functional brain networks [5], and to synthesize missing longitudinal scans.
References
[1] Zhao et al.: Variational Autoencoder For Regression: Application to Brain Aging Analysis, Medical Image Computing and Computer-Assisted Intervention, 2019
[2] Zhao, Liu et al.: Longitudinal Self-Supervised Learning, Medical Image Analysis, 2021
[3] Ouyang, Zhao et al.: Disentangling Normal Aging from Severity of Disease via Weak Supervision on Longitudinal MRI, IEEE TMI, 2022
[4] Ouyang et al.: Self-supervised Learning of Neighborhood Embedding for Longitudinal MRI, MEDIA, 2022
[5] Zhao et al.: Variational Autoencoder with Truncated Mixture of Gaussians for Functional Connectivity Analysis, Information Processing in Medical Imaging, 2019