Interpretable Deep Learning for Neuroimaging Studies
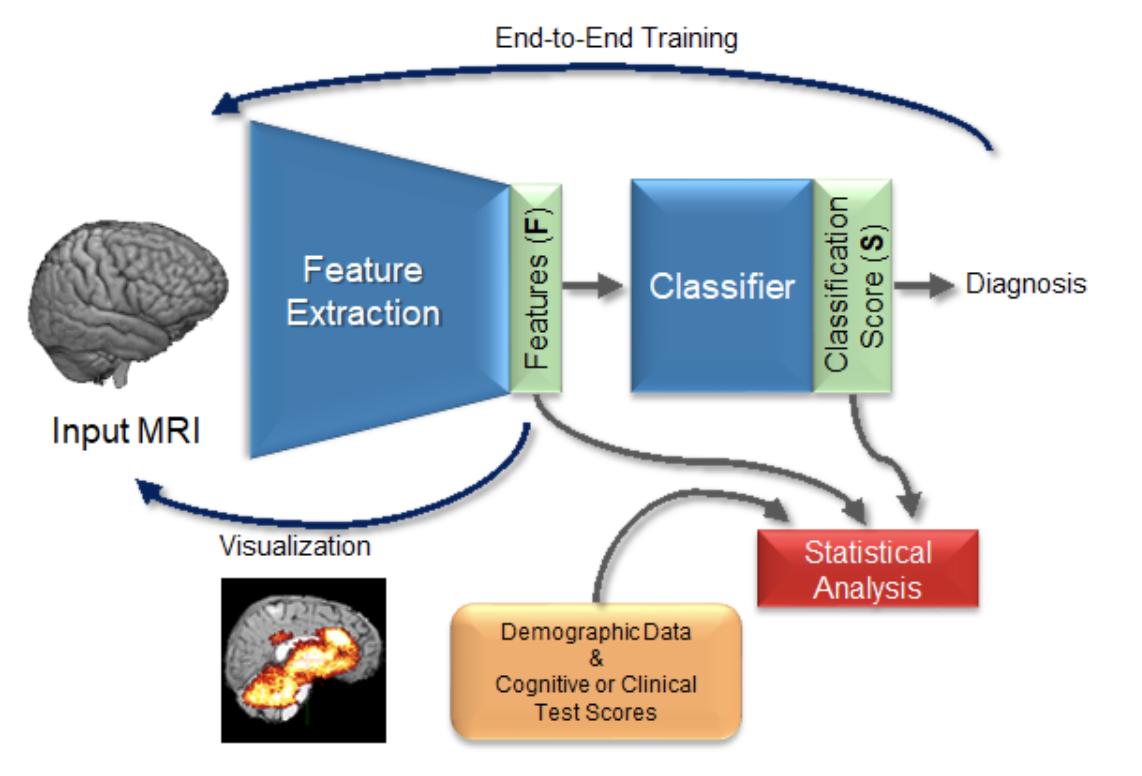
When machine learning methods are applied to neuroimaging and computational neuroscience applications, it is not only important to enable prediction of disease outcomes, but also to understand the underlying reasons why each subject is classified with any specific disease. Such an interpretation contribute to a mechanistic understanding of the diseases and may help clinicians design new therapeutic procedures. To this end, we conduct several research projects on the interpretability and explainability of deep learning methods applied to neuroimages.
As shown in computer vision, the power of deep learning lies in automatically learning relevant and powerful features for any perdition task, which is made possible through end-to-end architectures. However, deep learning approaches applied for classifying medical images do not adhere to this architecture as they rely on several pre- and post-processing steps. This shortcoming can be explained by the relatively small number of available labeled subjects, the high dimensionality of neuroimaging data, and difficulties in interpreting the results of deep learning methods. In a set of projects (ongoing and past, e.g., [1][2][7]), we proposed simple 3D Convolutional Neural Networks and exploit its model parameters to tailor the end-to-end architecture for the diagnosis of different diseases from MRIs. Based on the learned model, we identify the disease biomarkers and validate the results by exploring the learned features or brain regions, and relations to the clinical literature.
Another important factor in studying the brain is consideration of confounding factors in the studies. Presence of confounding effects is inarguably one of the most critical challenges in all medical applications. They influence both input (e.g., neuroimages) and the outcome (e.g., diagnosis or clinical score) variables and may cause spurious associations when not properly controlled for. Confounding effect removal is particularly difficult for a wide range of state-of-the-art prediction models, including deep learning methods. These methods operate directly on images and extract features in an end-to-end manner. This prohibits removing confounding effects by traditional statistical analysis, which often requires precomputed features (image measurements, like brain regional volumes). In these works, we study different methods to learn confounder-invariant discriminative features [3][4], conduct confounder-aware visualization of convolutional neural networks [5], and develop deep learning normalization operators that correct the unintended effects of confounders [6].
References
[1] Esmaeilzadeh et al. End-To-End Alzheimer’s Disease Diagnosis and Biomarker Identification, International Workshop on Machine Learning in Medical Imaging, 2018
[2] Adeli, Zhao, et al.: Deep learning identifies morphological determinants of sex differences in the pre-adolescent brain, NeuroImage, 2020
[3] Adeli, Zhao, et al.: Representation Learning with Statistical Independence to Mitigate Bias, IEEE/CVF Winter Conference on Applications of Computer Vision, 2021
[4] Zhao, Adeli, et al.: Training confounder-free deep learning models for medical applications, Nature Communications, 2020
[5] Zhao, Adeli, et al.: Confounder-Aware Visualization of ConvNets, International Workshop on Machine Learning and Medical Imaging, 2019
[6] Lu, et al.: Metadata Normalization, Conference on Computer Vision and Pattern Recognition, pp. 10917-10927, 2021
[7] Zhang, Zhao et al.: Multi-Label, Multi-Domain Learning Identifies Compounding Effects of HIV and Cognitive Impairment, MEDIA, 2022.