Deep Learning for Longitudinal Neuroimaging Data
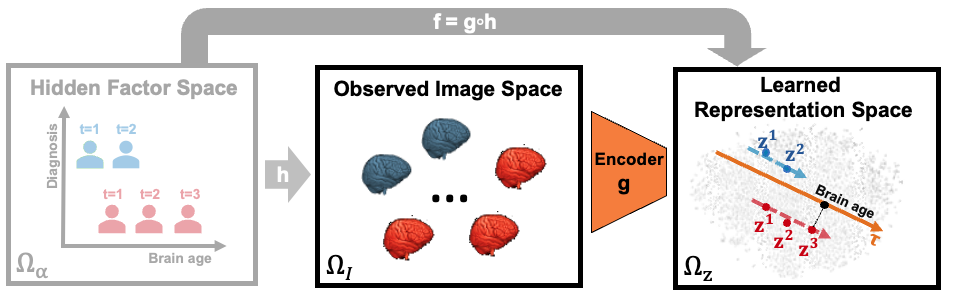
Longitudinal neuroimaging studies enable scientists to track the gradual effect of neurological diseases and environmental influences on the brain over time. To quantify those effects, we aim to enhance the applicability of deep learning to longitudinal MRI data by proposing both unsupervised and supervised models. To distinguish the longitudinal MRIs of healthy controls from those of the cohort of interest, existing deep learning models often couple Convolutional Neural Networks (CNN) with Recurrent Neural Networks (RNN), where the CNN reduces each MRI of the longitudinal sequence to informative features and the RNN uses the features to predict cohort assignment at each visit. We extend this CNN+RNN architecture with several mechanisms, including a novel longitudinal layer and consistency regularization, to model realistic disease progression along time. We also design self-supervised and unsupervised representation learning models based on parametric and non-parametric variational autoencoders (VAE) to allow the tracking of longitudinal effects in the features extracted from MRIs.
References
[1] Ouyang et al.: Longitudinal Pooling & Consistency Regularization to Model Disease Progression from MRIs, IEEE Journal of Biomedical and Health Informatics, 2021
[2] Zhao et al.: LSSL: Longitudinal Self-Supervised Learning, Medical Image Analysis, 2021
[3] Ouyang, Zhao et al.: Disentangling Normal Aging from Severity of Disease via Weak Supervision on Longitudinal MRI, IEEE TMI, 2022
[4] Ouyang et al.: Self-supervised Learning of Neighborhood Embedding for Longitudinal MRI, MEDIA, 2022
[5] Paschali et al.: Detecting Negative Valence Symptoms in Adolescents based on Longitudinal Self-Reports and Behavioral Assessments, JAD, 2022