Statistical and Machine Learning Models for Functional MRI Analysis
The Blood-Oxygen-Level-Dependent (BOLD) signal of rs-fMRI characterizes the intrinsic functional organization of the human brain by measuring its spontaneous activity at rest. Accurate analysis of rs-fMRI can enhance the understanding of functional neurodevelopment across the life span, characterize developmental disruption caused by neurological disorders, and explain differences in cognitive performance linked to sex.
To improve characterization of functional neurodevelopment, we have developed novel computational tools for longitudinal rs-fMRI data by deriving trajectories of functional networks that are biologically plausible [1][2]. We then improve the analysis of longitudinal trajectories of connectivity patterns by studying the manifold of positive-definite cone (that underlies the connectivity data) incorporating theories of parallel-transport and Lie group action [3]. These methods were shown to have improved statistical power in detecting group differences in functional development of the brain.
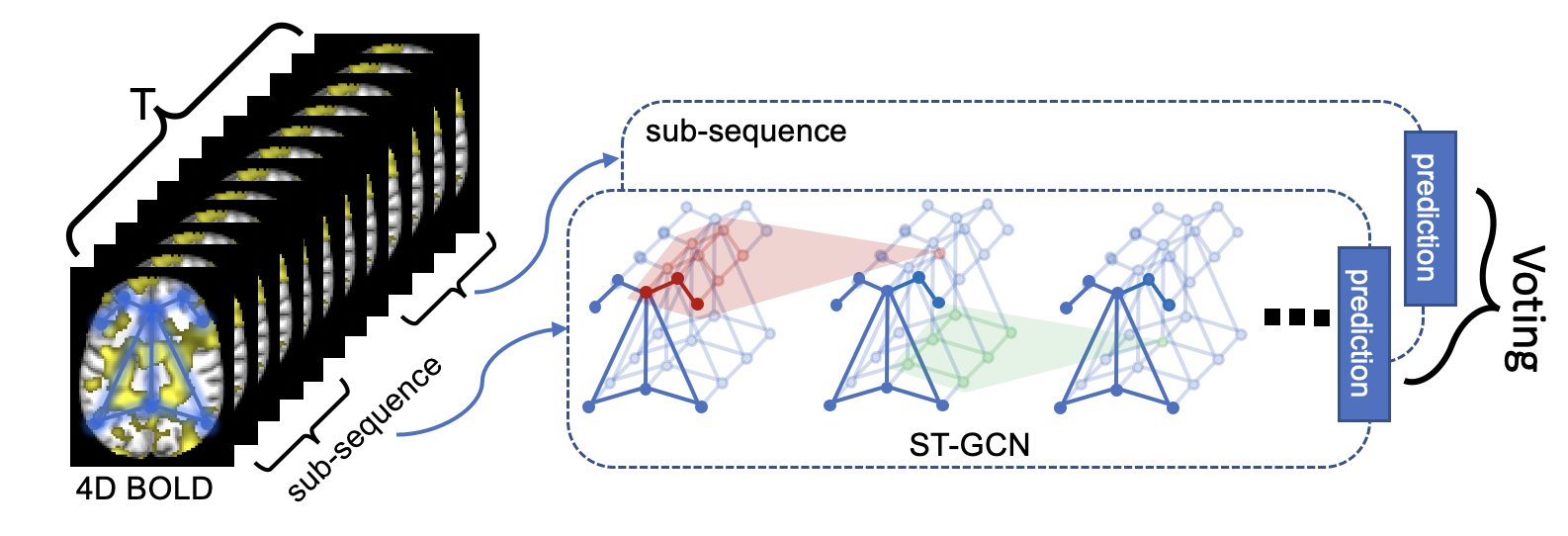
In addition to investigating the change of functional organization over a long period of time, we also study the meaningful variations within the time series captured by the single session of resting-state fMRI, i.e. dynamic functional connectivity. We do so by designing a computationally efficient model based on linear covariance shrinkage to compute large covariance matrices using very few time samples [4]. Major dynamic states are then estimated by clustering these covariance matrices, which is achieved by a novel unsupervised model called Truncated Gaussian-Mixture Variational AutoEncoder (TGM-VAE)[5]. Lastly, we also use deep learning to associate patterns of dynamic functional connectivity with key factors (disease, age, gender) by designing models specifically for the 4D spatio-temporal BOLD data [6], positive-definite correlation matrices [7], and coupling the functional connectivity with the underlying structural connectivity quantified by Diffusion Tensor Imaging .
References
[1] Zhao et al.: Longitudinally Consistent Estimates of Intrinsic Functional Networks, Human Brain Mapping, 2019
[2] Zhao et al.: Adolescent alcohol use disrupts functional neurodevelopment in sensation seeking girls, Addiction Biology, 2020
[3] Zhao et al.: A Riemannian Framework for Longitudinal Analysis of Resting-State Functional Connectivity, Medical Image Computing and Computer-Assisted Intervention, 2018
[4] Honnorat et al.: Covariance Shrinkage for Dynamic Functional Connectivity, Connectomics in Neuroimaging, 2019
[5] Zhao et al.: Variational Autoencoder with Truncated Mixture of Gaussians for Functional Connectivity Analysis, Information Processing in Medical Imaging, 2019
[6] Gadgil et al.: Spatio-Temporal Graph Convolution for Resting-State fMRI Analysis, Medical Image Computing and Computer-Assisted Intervention, 2020
[7] Honnorat et al.: Deep Parametric Mixtures for Modeling the Functional Connectome, Predictive Intelligence in Medicine, 2020
[8] Li et al.: Joint Graph Convolution for Analyzing Brain Structural and Functional Connectome, MICCAI 2022